 Fancy a part-share in the second most powerful supercomputer in the world? Think what you could do with it: play Call of Duty or Crysis faster than real-time, crack all your friends’ passwords in a microsecond, finish your accounts spreadsheets before you even started, forecast hurricanes, predict climate change…. If the mood took you, you could even model the behaviour and effects of biological molecules in the human body. Dream on – unless the last one appealed to you, in which case you can start straight after you’ve finished reading.
Fancy a part-share in the second most powerful supercomputer in the world? Think what you could do with it: play Call of Duty or Crysis faster than real-time, crack all your friends’ passwords in a microsecond, finish your accounts spreadsheets before you even started, forecast hurricanes, predict climate change…. If the mood took you, you could even model the behaviour and effects of biological molecules in the human body. Dream on – unless the last one appealed to you, in which case you can start straight after you’ve finished reading.
Supercomputers rely on parallel processing – joining together huge numbers of processors, each of which does a small part of the job at the same time as all the others. Think computer CPUs with thousands of cores instead of just the two, four or eight. There is no reason why all these processors need to be in the same place, indeed some could be in your PC. This is the idea behind Folding@home, a massive computing effort run by the School of Medicine at Stanford University in the USA, which gives medical researchers access to enormous amounts of distributed computing effort to do simulations of molecular dynamics. These are part of studies at the Stanford Pande Lab to try to work out the processes behind protein “folding” – proteins folding the wrong way are believed to be the causes of diseases like Alzheimer’s, Huntington’s chorea, BSE, CJD, Parkinson’s disease, and certain cancers. A worthy effort, therefore, but how do you take part?
You run the Folding software on your machine, and Stanford automatically allocates you “work units”, each of which can take up to around a week to complete. You don’t have to do anything – the software runs quietly in the background using up your spare CPU cycles, either while you are doing other things, or, if you need to use the full power of your PC, when it is idle and you are not using it at all (e.g. overnight). You can do this as an “Anonymous” contributor, or you can register a name, and set up or join a team of contributors. You will earn points for your contributions, although I’m not sure what you can do with them other than climb a league table – unfortunately I don’t think they get you a discount off brain surgery.
Currently there are 273,733 volunteered machines, making up the second biggest supercomputer in the world at 45,547 teraflops . (A teraflop is a thousand billion floating-point operations per second.) Number one is the Chinese MilkyWay-2 at the National Super Computer Centre in Guangzhou, at 54,902 teraflops, and number two, or three if you count Folding@home, is the Titan – Cray XK7 at Oak Ridge National Laboratory, at 17,590 teraflops. OK, maybe the numbers aren’t comparable quite as simply as this, but they give you an idea of the scale.
Folding@home has some impressive sponsors, in fact I first found out about it while installing an ATI graphics driver:
Start Folding!
Interested? Go to http://folding.stanford.edu/home/ , click on the download link where it says “Start Folding”, and follow the instructions to download and install. The current version is 7.4.4, and you can also download it for various flavours of Linux. In fact I run it on both a Windows 8.1 and an Ubuntu 14.04 Linux machine. During the Windows installation you can default or select the folder in which the work will be stored; I always create a special folder called StanfordFoldingData. The idea is that if you have to reinstall the software, you will be able to select this folder and the software will take the work up again where it left off.
When you have finished the installation and let the software start, it will start your browser and invite you to give your Folding account a name, and a team number if you want to join one. You also have the option to request a Folding passkey so that no-one can hack your account and pilfer your hard-earned points. If you install in Windows 7, you will find that the F@H screensaver is available; this is a display of your current molecule against a world map background, and shows progress on the current work unit. (It should also work on Windows 8, but you might have to search for FAHScreensaver.scr and manually copy it into C:\\Windows\\System32, or C:\\Windows\\SysWOW64, or both.)
In the F@HClient menu, you will find a link for F@HControl . This brings up a detailed interface for various settings, including how much of your CPU you want to give it (Light, Medium, or Full). There are plenty of other settings for you to investigate in Configure and Preferences. For instance, in Configure you can select a particular project to which you want to contribute, specify your contribution account name and team number, and request and enter a passkey. Preferences lets you specify an appearance theme for the Control GUI, among other things.
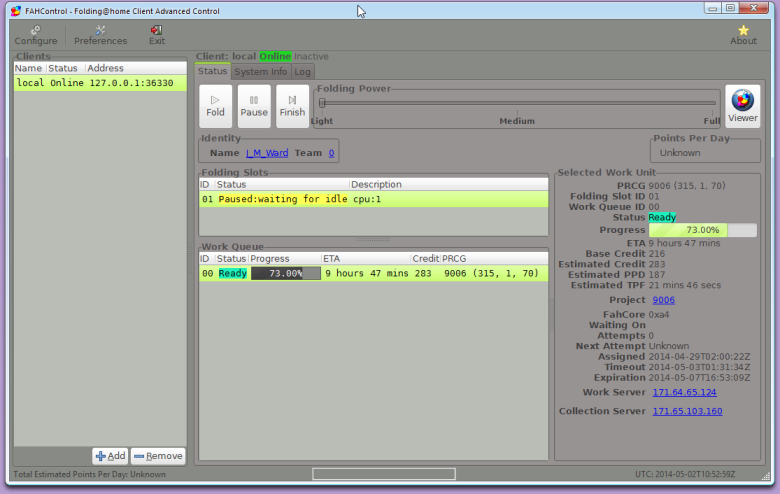
You can also change the CPU power you want to commit in the browser interface, where you can also select Idle, if you want F@H running only when you are not using your computer. As you can see, the browser screen tells you about progress on the current work unit, the number of points you’ve earned so far, and an outline of the project to which you are currently contributing.
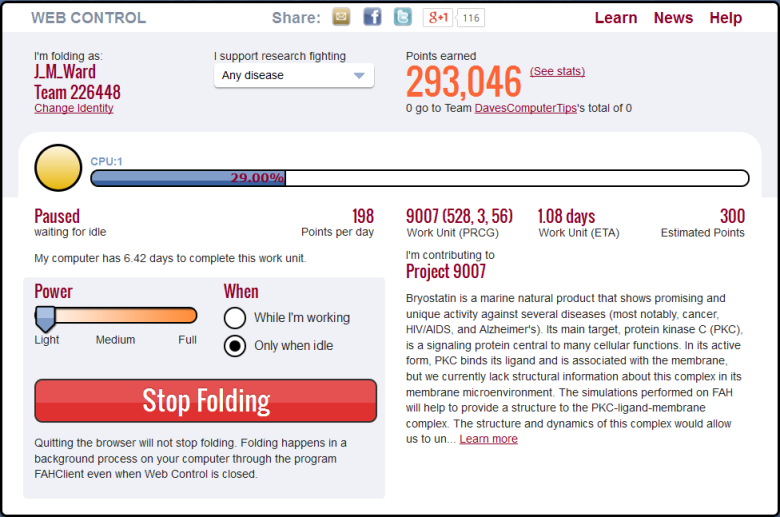
It’s best to let F@H run at the default settings unless you notice that your normal computer performance is adversely affected. If it is, then you can turn F@H down a notch, or go to the Idle setting.
From Control, you can start the Viewer, which will show you your molecule and computation progress; the screensaver has the same display.
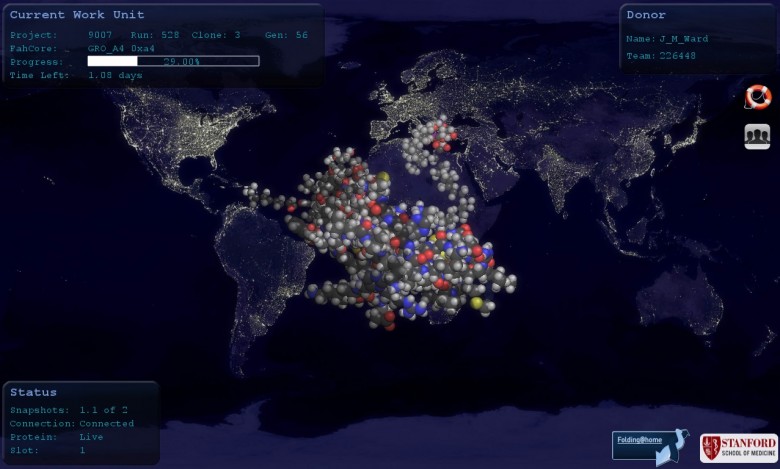
If you have a good modern graphics card, F@H will also set up a “slot” to use its GPU at the same time as the CPU. If this causes a problem, for example when you’re playing a game, you can just turn F@H down, as above. If it causes a serious problem which won’t go away, you can use Configure, with care, to remove the GPU slot altogether. My computer doesn’t have a suitable graphics card, so F@H hasn’t set up a GPU slot.
Join the Team!
Here at Daves Computer Tips, we have started an F@H team, called – guess what – DavesComputerTips. It would be great if you could join us, by specifying the team number, 226448, when you set up your software. Or later, you can select Change Identity on Web Control, or Configure | Identity in FAHControl. Then we can monitor progress under Team Stats on the Folding website, and see if we can get anywhere near the top of the league table. There’s some serious competition out there, people like Hewlett Packard and Overclockers Australia, with big GPUs and 8-core CPUs, who spend a lot of their time tuning up their systems to become giga-folders; still, it’s worth a try. Please join us!
Folding@home Team: DavesComputerTips
Team Number: 226448
You can find additional help in the Folding website installation instructions or FAQs, or on their forums – it’s well worth exploring the site to find out more about what they do with all that power. You can also ask questions here, and we’ll either answer directly or point you in the right direction.
P.S. – I lied about Call of Duty and Crysis – they won’t run on a supercomputer operating system, which needs to be super-reliable and super-efficient; that would be Linux.


Hi Martin,
I downloaded FAH, and am running it as we speak. I’m also adding points to the DCT Team.
Of course, when watching the Viewer, I have no idea what I’m looking at :-
Having fun with this,
Richard
Hi Richard,
Congratulations on becoming a founder member of the DavesComputerTips F@H team! You will see from the Web GUI image above that at the time I joined we had the grand total of zero points, but I hope that with your help and that of other DCT readers we will soon accelerate away from that situation!
As I understand it, what you are seeing in the Viewer (apart from the world map, which I hope you did recognise!) is a visualisation of the molecule that your CPU is helping to simulate. If you click on “Learn More” under the Project number in the Web GUI, or on the Project number at the right in F@H Control, you will get a brief description of the project to which you are contributing. You may need to reach for a biochemistry textbook to understand this. In my case (Project 9007), you also get a picture of the Ph.D. student doing the work, Steven Ryckbosch, holding a rabbit. Maybe the rabbit is part of the project – not sure, but if it is, maybe it won’t get Alzheimer’s. Which begs the question – how do you tell whether a rabbit has Alzheimer’s? I’m still working on that one.
Hi Martin,
Thanks for the info.
I got my first work unit completed today and the DCT score is up a bit now. I hope others join in!
You can tell that a rabbit has the disease when he forgets to progenate.
Certainly, I thought you knew that, 🙂
Richard
Hi Martin,
Where can I find the DCT Team list at FAH?
Thanks in advance,
Richard
Hi Richard,
If you click on Team Statistics on the F@H home page, you will get a ranked list of the teams. If you click on a team name, let’s say Overclock.net, you will get Overclock.net’s team page. At the bottom there is a Fast Teampage URL. If you click it you will get a list of the contributing team members. However, on the corresponding DavesComputerTips team page, there is no Fast Teampage URL listed. I suspect this is because the team has just formed and has very few members as yet. I will keep an eye on this, and if it hasn’t changed in the next month or so, I will enquire with the F@H administration people.
Sorry about the slightly delayed replies; I have spent all night reading Glenn Greenwald’s new book “No Place to Hide”, couldn’t put it down and now I’m starting over.
Hi Martin,
I found the link right on the Control Client under the Status Tab.
If you click on the Team ID number, it will open up in your browser and display our progress,
Richard
Excellent stuff Martin!
I’ll be getting on board any minute now.
Martin, a bit late, but I joined today.